Research Article | Open Access
Bioinformatics-based analysis of Cordyceps Sinensis in the treatment of diabetic nephropathy-associated depression
Xiaohui Li1, 2, 3, Yajun Qiao2, 3, Xingfang Zhang4, Ruiying Cheng3, Yi Liu1, Huimin Zheng5, Lixin Wei3, Yi Ding4, Hongtao Bi1, 3, Tingting Gao2
1School of Pharmacy, Qinghai University, Xining 810016, China.
2School of Psychology, Chengdu Medical College, Chengdu 610500, China.
3Qinghai Provincial Key Laboratory of Tibetan Medicine Pharmacology and Safety Evaluation, Northwest Institute of Plateau Biology, Chinese Academy of Science, Xining 810001, China.
4Department of Pharmacy, Xijing Hospital, Fourth Military Medical University, Xi’an, 710032, China.
5Department of Pharmacy, Qinghai Minzu University, Xining 810007, China.
Correspondence: Yi Ding (Department of Pharmacy, Xijing Hospital, Fourth Military Medical University, Xi’an, Shaanxi Province, 710032, China; E-mail: dingyi.007@163.com), Hongtao Bi (Qinghai Provincial Key Laboratory of Tibetan Medicine Pharmacology and Safety Evaluation, Northwest Institute of Plateau Biology, CAS, 23 Xinning Road, Xining 810008, China; Email: bihongtao@hotmail.com) and Tingting Gao (School of Psychology, Chengdu Medical College, 783 Xindu Road, Chengdu 610500, China; Email: gaott646@163.com).
Asia-Pacific Journal of Pharmacotherapy & Toxicology 2025, 5: 53-64. https://doi.org/10.32948/ajpt.2025.05.20
Received: 15 Jun 2025 | Accepted: 18 Sep 2025 | Published online: 10 Oct 2025
Methods Active components of CS were screened via TCMSP, disease-related targets were identified from GeneCards and OMIM, and potential targets were determined by intersection analysis. PPI network, GO/KEGG enrichment analyses, and "drug-component-target-pathway" network were constructed. Molecular docking and molecular dynamics simulations were employed to verify binding affinity and stability.
Results Seven active ingredients (e.g.β-sitosterol, ergosterol) and 40 key targets were identified, with PIK3CA, AKT1, and MAPK1 as core hubs. CS primarily modulated PI3K-Akt, MAPK-ERK, and androgen receptor pathways. Molecular docking revealed binding energies < -7.0 kcal/mol, and MD simulations confirmed complex stability within 50 ns.
Conclusion CS treats DN with depression via multi-component and multi-target synergism, targeting PIK3CA/AKT1/MAPK1 to regulate signaling pathways, thereby offering mechanistic molecular bases for clinical intervention.
Key words cordyceps sinensis, diabetic nephropathy, depression, network pharmacology, molecular docking, molecular dynamics simulation
Depression is a chronic, relapsing mental disorder marked by persistent low mood, cognitive slowing, poor concentration, anhedonia, and loss of interest and is often accompanied by somatic symptoms including sleep disturbances and appetite loss [4-6]. The prevalence of depression is currently increasing dramatically worldwide, with a prevalence rate of 4.4% [7]. There is a complex interaction between diabetes and depression. Dysregulation of the hypothalamic‒pituitary‒adrenal‒immune (HPAI) axis and activation of proinflammatory cytokines are associated with depression, which may lead to insulin resistance and increase the risk of diabetes mellitus [8].
Cordyceps sinensis (CS), a renowned traditional Chinese medicine, is considered a valuable natural resource and cultural heritage of China [4]. It represents a unique parasitic complex formed by the fusion of cysts of the CS and the corpse of the larvae. The Cordyceps fungus belongs to the family Clavicipitaceae [9]; it parasitizes the body of larvae of the family Batidae. Naturally, CS thrives in high-altitude regions, typically between 3,500 and 5,000 meters above sea level, such as Tibet, Qinghai, Gansu, Sichuan, and Guizhou, where the environmental conditions favor its growth and development [10]. In addition to its significant immunomodulatory [11], antioxidative stress [12], and anti-inflammatory pharmacological activities, CS can improve renal function and protect the kidneys from damage by alleviating glomerulosclerosis and fibrosis, and by inhibiting the inflammatory response and oxidative stress. Moreover, CS has also shown potential efficacy in improving depressive symptoms by regulating the central nervous system, intestinal flora [13], and immune function. Therefore, CS has unique advantages in the treatment of depression complicated by diabetic nephropathy.
Currently, for the treatment of depression complicated by DN, Western medical therapies have limited effects. Although antidepressants like selective 5-hydroxytryptamine reuptake inhibitors (SSRIs) and tricyclic antidepressants (TCAs) can alleviate depressive symptoms, they have limited therapeutic effects on DN and may aggravate the burden on renal function [14, 15]. In addition, these drugs have many side effects, and their long-term use may lead to metabolic disorders and increased cardiovascular risk. Therefore, developing a multitarget therapeutic regimen that improves renal function and relieves depressive symptoms is imperative.
However, the specific mechanism of action of CS as a treatment for depression complicated by DN has not yet been clarified. Here, we employ network pharmacology along with complementary analytical approaches to investigate the underlying mechanisms through which CS serves as a treatment of depression complicated by DN, aiming to lay a theoretical foundation for subsequent studies. Figure 1 represents the research flowchart.
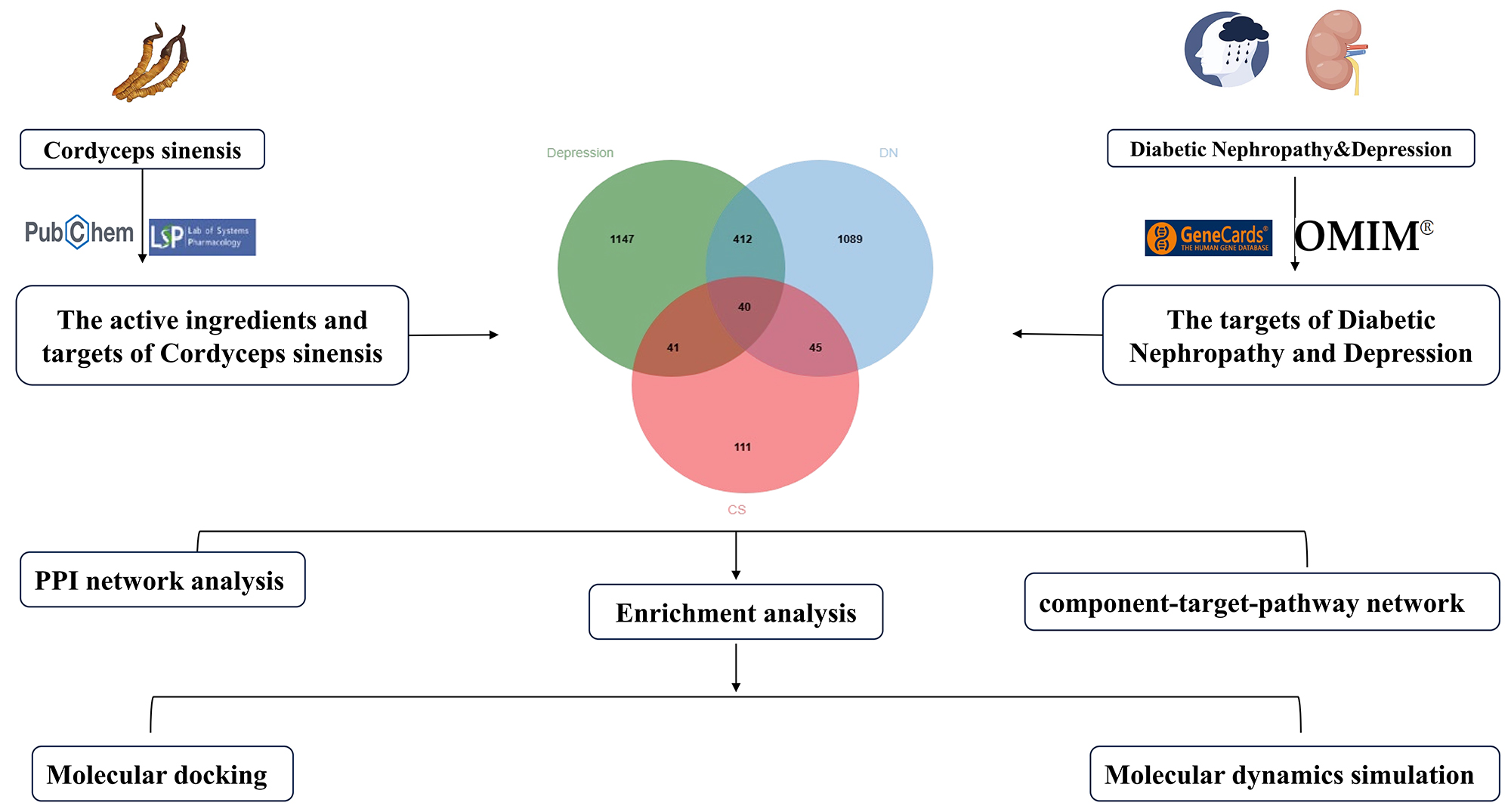 Figure 1. The flow chart of this study.
Figure 1. The flow chart of this study.
The constituents of CS were obtained from the Traditional Chinese Medicine System Pharmacology Database and Analysis Platform (TCMSP) (https://old.tcmsp-e.com/tcmsp.php) [16]. According to previous literature, compounds with oral bioavailability (OB) ≥ 30% and drug-likeness (DL) ≥ 0.18 were selected as screening criteria to identify the orally active components of CS.
Prediction of the action targets of CS active ingredients
The screened active components were retrieved from PubChem (https://pubchem.ncbi.nlm.nih.gov/) to obtain their corresponding SMILES structures [17]. These SMILES were then imported into the SwissTargetPrediction database (http://www.swisstargetprediction.ch/) database [18], with the target species restricted to humans (Homo sapiens). Targets with a predicted probability ≥ 0 were selected as the screening criterion, and the most relevant potential target information for each active compound was subsequently obtained.
Prescreening for disease target acquisition
Using “Depression” and “Diabetic Nephropathy” as keywords, we searched the GeneCards (https://www.genecards.org/) [19] and OMIM (https://www.genecards.org/) [20] databases for disease targets. The OMIM database was used to search for disease targets to screen the targets of DN and depression and removes duplicates from the two databases before organizing and merging the information to obtain the disease targets related to depression and DN. Using the jvenn (https://jvenn.toulouse.inrae.fr/app/example.html) platform, the intersection between the disease-related targets and the predicted targets of the active ingredients was identified, yielding the potential therapeutic targets of CS for depression and DN.
Prediction of protein‒protein interaction (PPI) information
The intersecting targets were uploaded into the STRING (https://cn.string-db.org/) [21] database, and on the search page, “Multiple proteins” and “Homo sapines” were selected for each species. The unconnected targets were removed to obtain the PPI network diagram.
Gene Ontology (GO) analysis
The shared targets of CS for the treatment of DN complicated with depression were uploaded to the DAVID (https://davidbioinformatics.nih.gov/) [22] database for GO enrichment analysis. The results from three items, biological process (BP), cellular component (CC), and molecular function (MF), were visualized and displayed via Bioinformatics (http://www.bioinformatics.com.cn/).
Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis
KEGG pathway analysis was performed via the DAVID database for common targets, and the results were visualized via Bioinformatics. (http://www.bioinformatics.com.cn/).
Construction of a “drug‒component‒target‒pathway” network
An Excel file of “drug-ingredient-target-pathway” was created with the active ingredients of CS, the intersecting targets of CS in the treatment of DN complicated with depression, and the pathways obtained from the enrichment analysis, which were then imported into Cytoscape 3.10 to construct a network diagram showing the interactions between the four components.
Molecular docking
The three core target proteins screened were used to perform molecular docking with the receptor proteins of their active ingredients. The 3D structures of the core target and protein receptor were obtained from the PubChem, PDB [23], and UniProt databases [24], and molecular docking was carried out via AutoDock vina1.5.7 software to determine the interaction strengths of the core target and the active ingredient. The docking interactions were visualized using PyMOL software, while Origin software was employed to generate thermograms illustrating the molecular docking binding energies.
Molecular dynamics simulation
The molecular dynamics (MD) simulations were carried out in the Yinfo Cloud Computing Platform (YCCP) via the Amber Tools 20 package with AMBER ff19SB [25] and GAFF [26] force fields for the three groups of complexes with the highest binding affinities during the molecular docking process. The system was solvated via the TIP3P water model with a truncated octahedral water box with a 10 Å edge. Periodic boundary conditions (PBC) were applied to neutralize the system’s net charge using Na⁺ counterions. To eliminate unfavorable atomic contacts, two consecutive minimization steps were conducted—10,000 steps of steepest descent followed by 10,000 steps of conjugate gradient minimization. After energy minimization, the system underwent equilibration under NVT and NPT ensembles for 200 ps and 500 ps, respectively. The temperature was maintained at 300 K using a Berendsen thermostat with a 1 ps coupling constant, while the pressure was controlled at 1 atm using a Monte Carlo barostat with a 1 ps relaxation time. Subsequently, a 50 ns MD production run was performed under the NVT ensemble without any positional restraints. The resulting trajectories were analyzed using the CPPTRAJ module [27]. Finally, binding free energies were computed using the Molecular Mechanics Generalized Born Surface Area (MM/GBSA) method implemented in AmberTools 20, based on 200 snapshots extracted from the MD trajectory [28]. For each snapshot, the free energy of the receptor, ligand, and complex was calculated using a “single-trajectory” approach.
Thirty-eight components of CS were included in the TCMSP, and seven active components were obtained by screening with OB ≥ 30% and DL ≥ 0.18, as shown in Table 1. After screening via the Swiss Target Prediction database with probability>0 as the screening criterion, 237 targets corresponding to active ingredients were finally obtained after de-weighing.
Prediction of disease targets
Depression and DN-related targets were collected from the GeneCards and OMIM disease databases. As a result, 1586 targets for depression and 1640 targets for CS were obtained. A total of 1586 targets and 1640 targets for DN and the active targets of CS intersected with the other two diseases, and 85 targets intersected between depression and CS. Eighty-one targets intersected between DN and CS, and a total of 40 targets were obtained, as demonstrated by the visualized Venn diagram shown in Figure 2A.
PPI network construction
Forty common targets of CS for the co-treatment of DN and depression were uploaded to the STRING database, and the target protein PPI network was obtained, which was later visualized and analyzed via Cytoscape 3.10.0 software (Figure 2B). Overall, the network had a 40 nodes and 320 edges in total, with the average node degree value of 16.
Target enrichment analysis (GO analysis)
The 40 intersecting targets of CS for the treatment of DN and depression were subjected to GO enrichment analysis. A total of 341 GO entries were analyzed (Figure 3A-D), of which 263 entries were for BP, as shown in Figure 3A; 31 entries were for CC, as shown in Figure 3B; 48 entries were for MF, as shown in Figure 3C; and the top 10 entries with the smallest P values were visualized through bubble diagrams. The first 10 entries with the smallest P values among the entries were drawn into bubble diagrams for visualization, and the results suggested that the BPs were focused mainly on the positive regulation of transcription by RNA polymerase II, the positive regulation of gene expression, the negative regulation of gene expression, etc. The cellular components were mostly composed of the cytoplasm, cytoplasmic solute, nucleus, etc. The MFs were composed mainly of the functions of binding of the same proteins, binding of zinc ions, and binding of enzymes.
Target enrichment pathway analysis (KEGG analysis)
Pathway analysis of the intersection targets of disease and drug components was performed through the DIVAD database. A total of 135 pathways were obtained, and the top 20 items with the smallest P value were selected for graphical visualization, as shown in Figure 3E-F. The results of the analysis suggested that CS cotreats DN and depression via PI3K‒Akt, MAPK‒ERK, androgen receptor signaling, and other pathways.
Drug‒component‒target‒pathway network
The CS-constituent-target-pathway network was constructed via Cytoscape 3.10.1 software (Figure 4). The red triangle represents CS, the surrounding light pink color represents the core constituents of CS, the green rectangle at the bottom represents the targets of CS in treating the disease, and the blue color at the top represents the signaling pathways. This network contains 264 nodes (drug: 1; ingredient: 6; target: 237; pathway: 20) and 439 edges (drug-ingredient: 6; ingredient-target: 273; target-pathway: 196). The results of the analysis conducted through the network indicated that the top three targets with highest degree value were PIK3CA, AKT1, and MAPK1, which were taken as the core targets in this study for subsequent research.
Molecular docking
The top three ranked targets were screened for molecular docking with the top six core components via AutoDock Vina 1.5.7. A lower affinity (kcal/mol) value indicates more stable binding between the compound and target protein, reflecting stronger binding affinity between the two molecules. It is generally believed that a binding energy value below -4.25 kcal/mol indicates a certain binding activity between the compound and target protein; a value below -5.0 kcal/mol indicates better binding activity, and a value below -7.0 kcal/mol indicates particularly strong binding activity. The results, as shown in Table 2 and Figure 5A below, revealed that PIK3CA, AKT1, MAPK1 and the first six active ingredients all had good binding activities, with the highest binding activity of the PIK3CA protein to β-sitosterol reaching -9.4 kcal/mol, followed by the binding activity of the AKT1 protein to cerevisterol, with a binding activity of -9.2 kcal/mol, and the binding activity of the MAPK1 protein with cerevisterol was in third place, with a binding activity of -8.6 kcal/mol; thus, we can infer that PIK3CA may present a key target of CS for the treatment of DN and depression. As shown in Figure 5B, some molecular docking results are visualized, and all three groups of complexes clearly formed hydrogen bonds during the redocking process.
Molecular dynamics simulation
Three groups of complexes with the highest binding activity from the molecular docking experiments were screened for verification via MD simulations. System stability was monitored via root mean square deviation (RMSD), with lower average RMSD values reflecting greater conformational stability of the complexes throughout the simulation. According to Figure 6A, the RMSD values of the three sets of complexes remained low and smooth throughout the 50 ns of the simulation. These findings indicate that the ligands and receptors are more tightly bound and that the complexes are more stable. The root mean square fluctuation (RMSF) can characterize the fluctuation of small-molecule ligands on the spatial structure of proteins, and a larger value suggests that the residues of the proteins interact more heavily with small molecules. As shown in Figure 6B, the RMSF values of the three groups of complexes were greater during the simulation process, and the analysis revealed that TYR, MET, and ARG may serve as key residues in the binding of the AKT1–cerevisterol complex; GLY, VAL, and ASP may play critical roles in the MAPK1–cerevisterol interaction; and ARG, MET, and LYS are likely essential residues contributing to the stability of the PIK3CA-β-sitosterol complex. The radius of gyration (ROG) can characterize the spatial compactness and changes in the morphology of a molecular system during a simulation. According to the analysis of Figure 6C, the AKT1-cerevisterol complex has a small ROG of approximately 1.47 nm during the simulation process, which indicates that the structure of the complex is relatively compact, the complex is relatively stable, and the possibility of structural changes during the simulation process is relatively small. The complex MAPK1-cerevisterol maintained the ROG at approximately 2.18 nm during the simulation process, indicating that there is stability in the structure of the complex and that at the same time, there is space for flexibility and dynamic changes, and different degrees of conformational changes may occur during the simulation process. The ROG of the complex PIK3CA-β-sitosterol remains at approximately 3.3 nm, indicating that the complex structure is more flexible and less stable and is prone to conformational changes during the simulation process.
The MM/GBSA method is a well-established computational approach used to calculate the binding free energies following simulations to predict the stability of a complex. Binding free energies were calculated for 200-frame snapshots of MD trajectories via the MM/GBSA method in AmberTools 20. GGAS denotes the gas-phase total free energy, which is the sum of the van der Waals force (VDW) and electrostatic potential energy (EEL), and ΔG Solvation Free Energy (GSOLV) denotes the solvation energy, representing the sum of the nonpolar solvation energy (NOPOLAR) and the polar solvation energy (POLAR). The binding free energies of the three groups of complexes shown in Figure 6D are -21.22 kcal/mol, -35.22 kcal/mol, and -35.44 kcal/mol, respectively, where the van der Waals forces are -27.82 kcal/mol, -48.52 kcal/mol, and -60.5 kcal/mol, the electrostatic potential energies are -12.84 kcal/mol, -0.59 kcal/mol, and -13.53 kcal/mol, the nonpolar solvation energies are -3.17 kcal/mol, -5.89 kcal/mol, and -5.37 kcal/mol, all three of which are favorable for the binding of the three groups of small-molecule compounds to proteins, and electrostatic potential and the van der Waals energies are stronger and play a dominant role, whereas the nonpolar solvation energies are weaker and play a secondary role, and the polar solvation energies are unfavorable for the interaction between the two.
|
Table 1. The main ingredients of CS. |
|||
|
MOL number |
Compound name |
OB/% |
DL |
|
MOL001439 |
Arachidonic acid |
45.57 |
0.2 |
|
MOL001645 |
Linoleoyl acetate |
42.1 |
0.2 |
|
MOL000358 |
β-sitosterol |
36.91 |
0.75 |
|
MOL011169 |
Peroxyergosterol |
44.39 |
0.82 |
|
MOL008998 |
Cerevisterol |
39.52 |
0.77 |
|
MOL008999 |
Cholesteryl palmitate |
31.05 |
0.45 |
|
MOL000953 |
Cholesterol |
37.87 |
0.68 |
|
CS: cordyceps sinensis; DL: drug-likeness; OB: oral bioavailability. |
|||
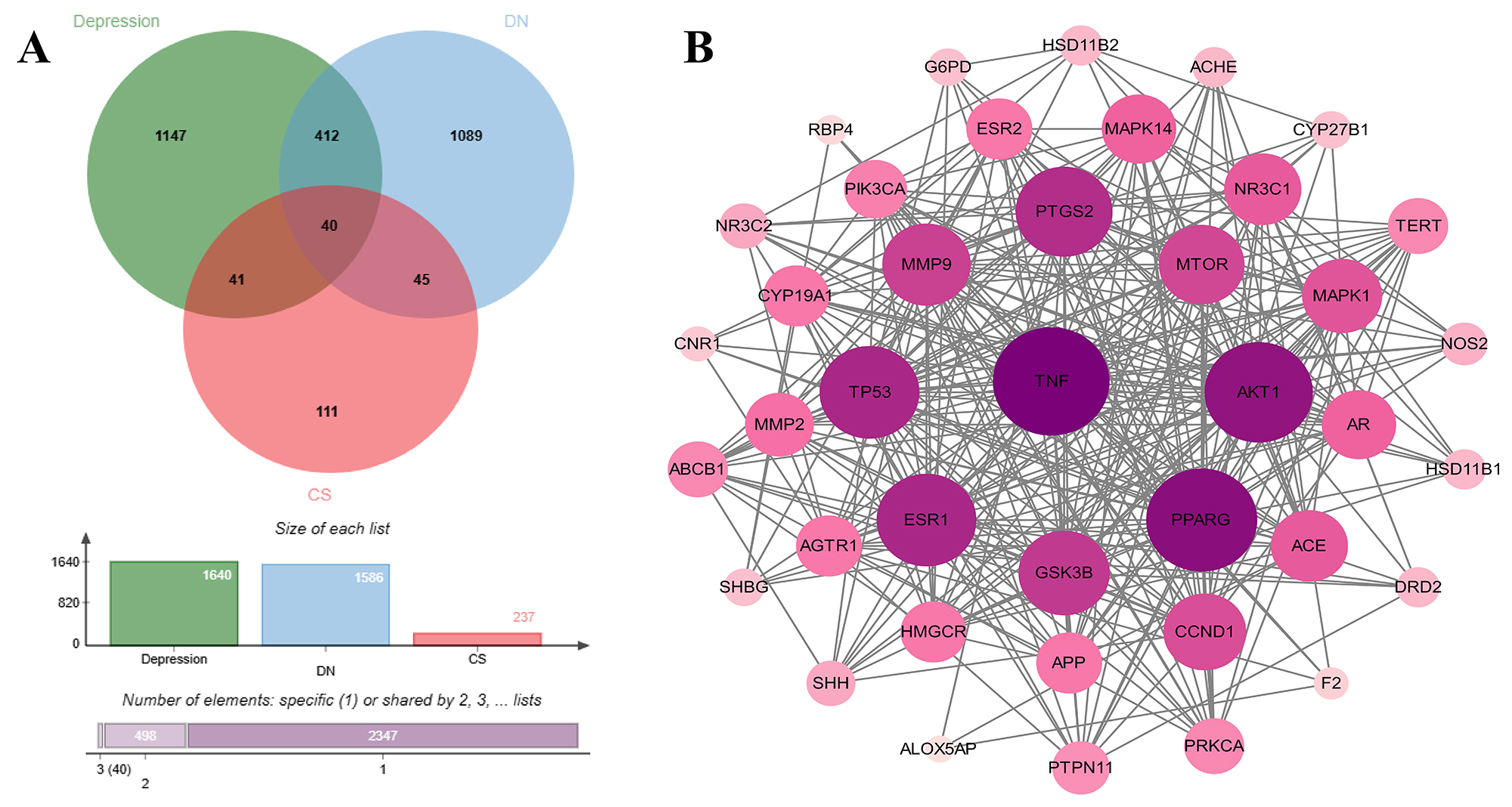 Figure 2. Analysis results of common targets and their interactions. (A) Effects of CS on the intersection of diabetic nephropathy and depression; (B) Protein‒protein network interaction map.
Figure 2. Analysis results of common targets and their interactions. (A) Effects of CS on the intersection of diabetic nephropathy and depression; (B) Protein‒protein network interaction map.
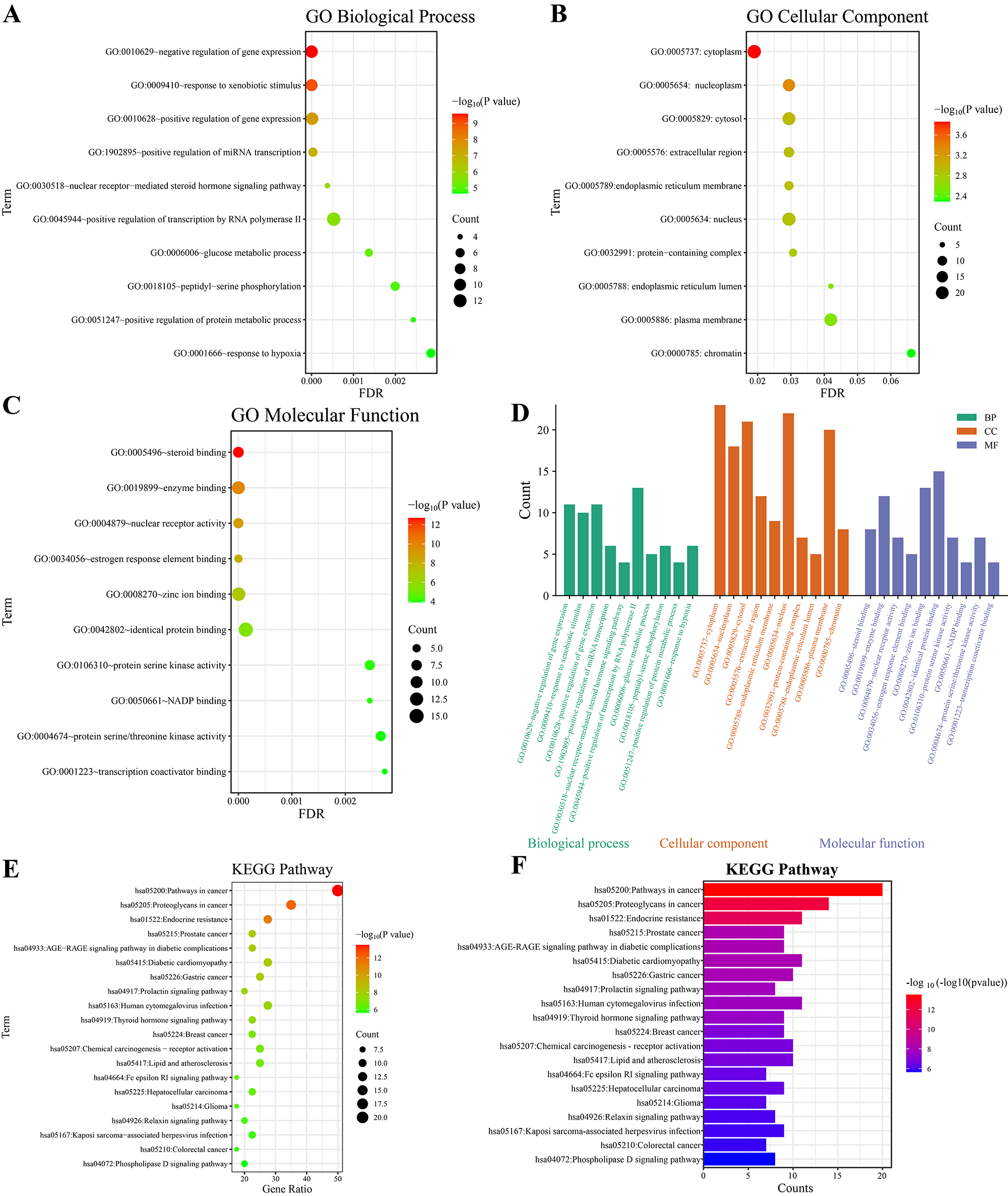 Figure 3. Enrichment analysis results. (A) GO BP enrichment analysis bubble map; (B) GO CC enrichment analysis bubble map; (C) GO MF enrichment analysis bubble map; (D) Histogram of the results of GO enrichment analysis; (E) Bubble diagram of the KEGG pathway enrichment analysis results; (F) KEGG pathway classification diagram.
Figure 3. Enrichment analysis results. (A) GO BP enrichment analysis bubble map; (B) GO CC enrichment analysis bubble map; (C) GO MF enrichment analysis bubble map; (D) Histogram of the results of GO enrichment analysis; (E) Bubble diagram of the KEGG pathway enrichment analysis results; (F) KEGG pathway classification diagram.
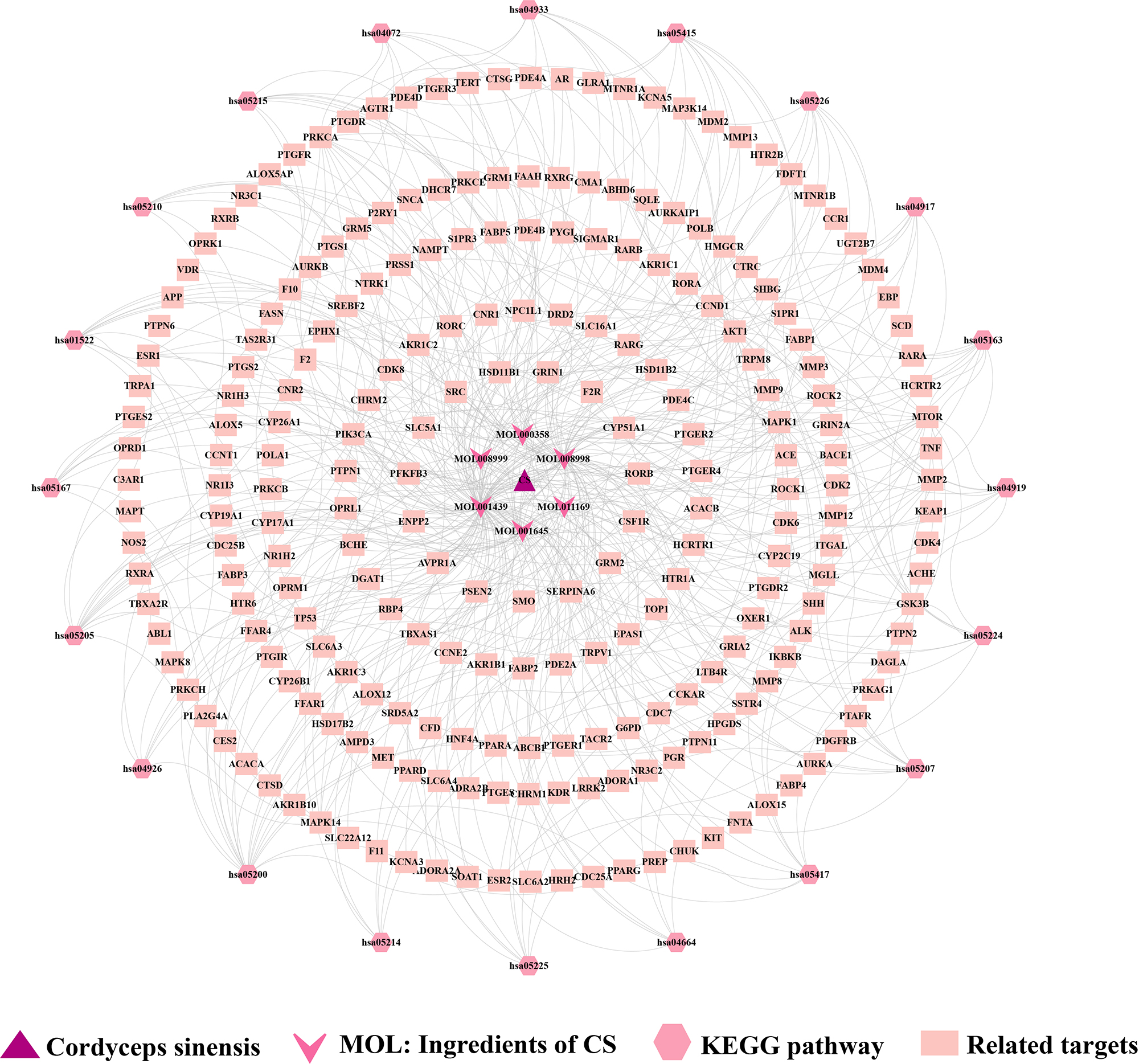 Figure 4. CS-component-target-pathway network diagram.
Figure 4. CS-component-target-pathway network diagram.
|
Table 2. Molecular docking results. |
|||||
|
Gene name |
Degree |
UniProt ID |
PDB ID |
Structural unit |
Binding energy (kcal/mol) |
|
PIK3CA |
21 |
P42336 |
2RD0 |
MOL001439 |
-6.4 |
|
|
|
|
|
MOL001645 |
-5.7 |
|
|
|
|
|
MOL000358 |
-9.4 |
|
|
|
|
|
MOL011169 |
-8.3 |
|
|
|
|
|
MOL008998 |
-8.2 |
|
|
|
|
|
MOL008999 |
-7.4 |
|
AKT1 |
21 |
P31749 |
1H10 |
MOL001439 |
-4.3 |
|
|
|
|
|
MOL001645 |
-4.2 |
|
|
|
|
|
MOL000358 |
-7.1 |
|
|
|
|
|
MOL011169 |
-7.4 |
|
|
|
|
|
MOL008998 |
-9.2 |
|
|
|
|
|
MOL008999 |
-5.1 |
|
MAPK1 |
20 |
P28482 |
1PME |
MOL001439 |
-5.4 |
|
|
|
|
|
MOL001645 |
-5.3 |
|
|
|
|
|
MOL000358 |
-8.1 |
|
|
|
|
|
MOL011169 |
-8.2 |
|
|
|
|
|
MOL008998 |
-8.6 |
|
|
|
|
|
MOL008999 |
-7.1 |
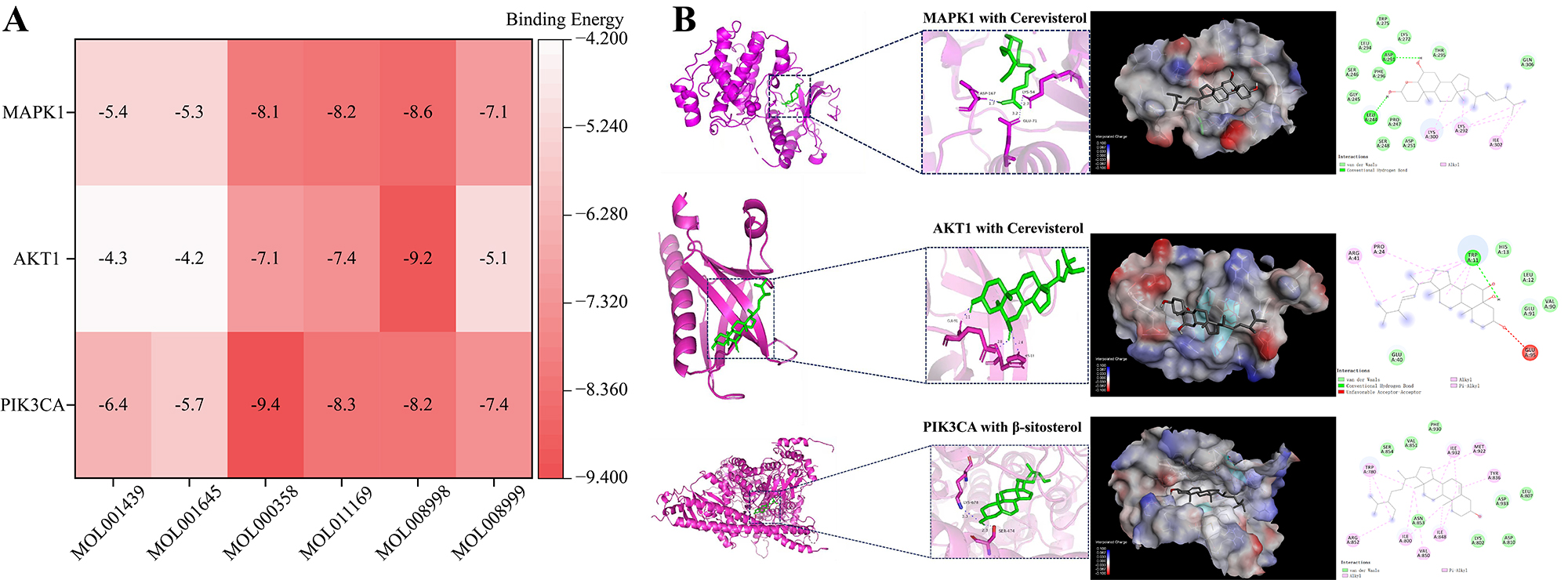 Figure 5. Results of Molecular docking. (A) Molecular docking binding energy heatmap (the color shading is inversely proportional to the magnitude of the binding energy); (B) Results of docking visualization of active ingredients with core target proteins.
Figure 5. Results of Molecular docking. (A) Molecular docking binding energy heatmap (the color shading is inversely proportional to the magnitude of the binding energy); (B) Results of docking visualization of active ingredients with core target proteins.
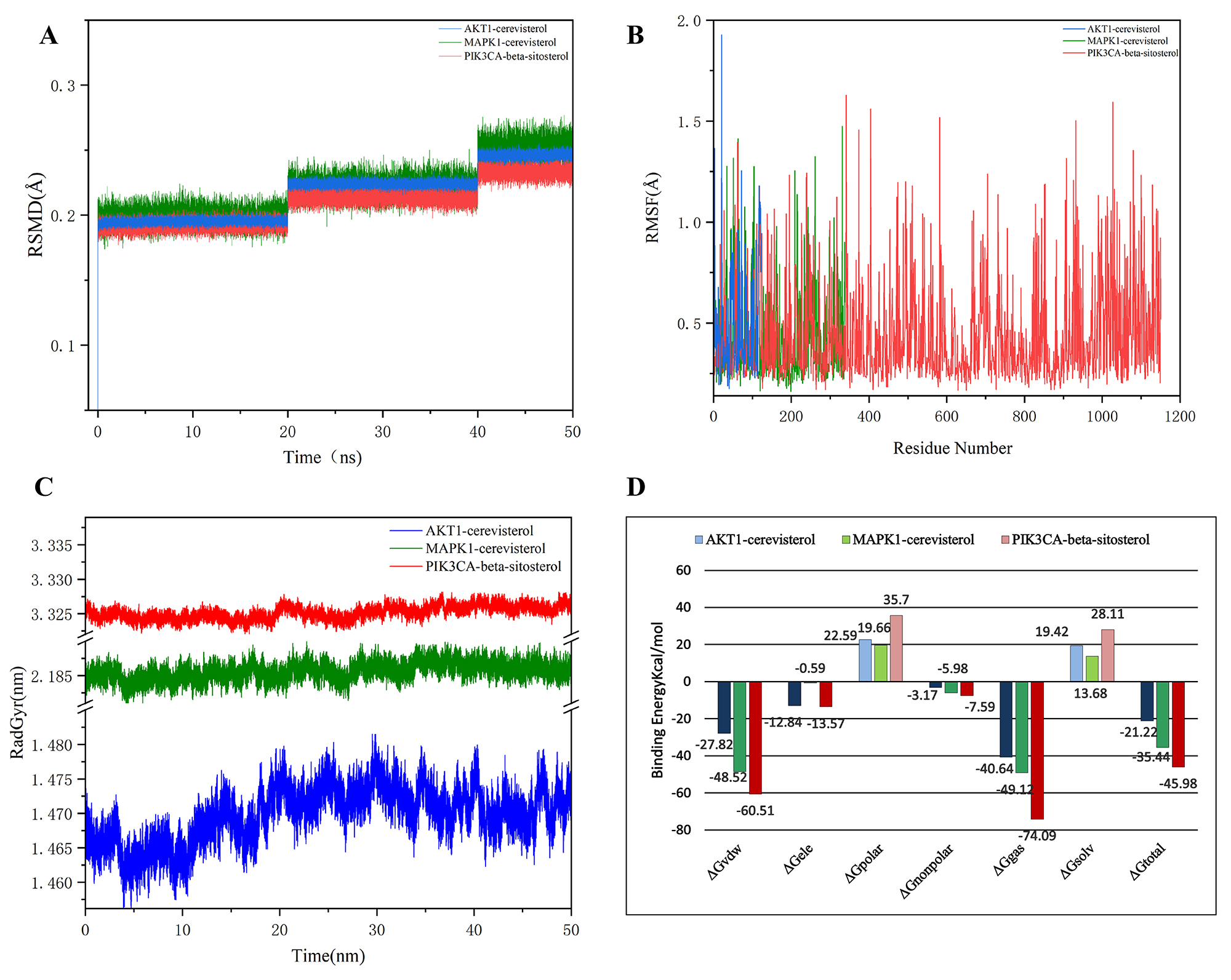 Figure 6. Results of molecular dynamics simulations. (A) RMSD curves of three complexes during molecular dynamics simulations; (B) RMSF curves of three complexes during molecular dynamics simulations; (C) ROG curves of three complexes during molecular dynamics simulations; (D) Molecular dynamics simulation energy analysis.
Figure 6. Results of molecular dynamics simulations. (A) RMSD curves of three complexes during molecular dynamics simulations; (B) RMSF curves of three complexes during molecular dynamics simulations; (C) ROG curves of three complexes during molecular dynamics simulations; (D) Molecular dynamics simulation energy analysis.
No applicable.
Ethics approval
No applicable.
Data availability
The data will be available upon request.
Funding
This work was supported by the Natural Science Foundation of China (Grant No. 82171863), the Innovation Platform Program of Qinghai Province (2021-ZJ-T02) and the Special Funds for the Central Government to Guide Local Scientific and Technological Development (2025ZY010).
Authors’ contribution
All authors have materially participated in the research and article preparation. The roles for all authors are as follows: Xiaohui Li: Designed the study, Data curation, Formal analysis, Writing—original draft; Yajun Qiao: Formal analysis, Investigation; Xingfang Zhang: Formal analysis, Investigation; Ruiying Cheng: Formal analysis; Yi Liu: Formal analysis; Huimin Zheng: Formal analysis; Lixin Wei: Formal analysis; Yi Ding: Formal analysis, Writing—review & editing; Hongtao Bi: Formal analysis, Writing—review & editing; Tingting Gao: Conceptualization, Writing—review & editing; All authors have approved the final version of the manuscript.
Competing interests
The authors declare that they have no competing interests.
- Shi L, Song Z, Li Y, Huang J, Zhao F, Luo Y, Wang J, Deng F, Shadekejiang H, Zhang M et al: MiR-20a-5p alleviates kidney ischemia/reperfusion injury by targeting ACSL4-dependent ferroptosis. Am J Transplant 2023, 23(1): 11-25.
- Wang Z, Fu W, Huo M, He B, Liu Y, Tian L, Li W, Zhou Z, Wang B, Xia J et al: Spatial-resolved metabolomics reveals tissue-specific metabolic reprogramming in diabetic nephropathy by using mass spectrometry imaging. Acta Pharm Sin B 2021, 11(11): 3665-3677.
- Kang JS, Cho NJ, Lee SW, Lee JG, Lee JH, Yi J, Choi MS, Park S, Gil HW, Oh JC et al: RIPK3 causes mitochondrial dysfunction and albuminuria in diabetic podocytopathy through PGAM5-Drp1 signaling. Metabolism 2024, 159: 155982.
- Zhang M, Li A, Yang Q, Li J, Zheng L, Wang G, Sun Y, Huang Y, Zhang M, Song Z et al: Matrine alleviates depressive-like behaviors via modulating microbiota-gut-brain axis in CUMS-induced mice. J Transl Med 2023, 21(1): 145.
- Kwon H, Seung H, Tran K, Yang I, Kim S: Efficacy and mechanism of Chinese herbal medicine Banxia-Houpo-Tang for depression: a meta-analysis and network pharmacology analysis. Tradit Med Res 2025, 10(5): 26.
- Mao J, Chen H, Wang M: Role of echinacoside in alleviating depressive-like symptoms in chronic unpredictable mild stressed mice via PI3K/AKT/Nrf2/HO-1 signaling pathway. Tradit Med Res 2026, 11(5): 40.
- Marwaha S, Palmer E, Suppes T, Cons E, Young AH, Upthegrove R: Novel and emerging treatments for major depression. Lancet 2023, 401(10371): 141-153.
- Xie Y, Bowe B, Mokdad AH, Xian H, Yan Y, Li T, Maddukuri G, Tsai CY, Floyd T, Al-Aly Z: Analysis of the Global Burden of Disease study highlights the global, regional, and national trends of chronic kidney disease epidemiology from 1990 to 2016. Kidney Int 2018, 94(3): 567-581.
- Zhang X, Qiao Y, Wang M, Liang X, Zhang M, Li C, Cairang J, Wang J, Bi H, Gao T: The influence of genetic and acquired factors on the vulnerability to develop depression: a review. Biosci Rep 2023, 43(5): BSR20222644.
- Becker LJ, Fillinger C, Waegaert R, Journée SH, Hener P, Ayazgok B, Humo M, Karatas M, Thouaye M, Gaikwad M et al: The basolateral amygdala-anterior cingulate pathway contributes to depression-like behaviors and comorbidity with chronic pain behaviors in male mice. Nat Commun 2023, 14(1): 2198.
- Lu H, Shen M, Chen Y, Yu Q, Chen T, Xie J: Alleviative effects of natural plant polysaccharides against DSS-induced ulcerative colitis via inhibiting inflammation and modulating gut microbiota. Food Res Int 2023, 167: 112630.
- Jiang Y, Liao Y, Liu Z, Zhou M, Wang H, Qi H, Sun S, Xi S, Tang Y: The effects of Cordyceps polysaccharides on ischemic brain injury in rats via intervening with IL-23/IL-17 axis and the intestinal barrier. Int J Biol Macromol 2024, 283(Pt 2): 137526.
- Ji Y, Tao T, Zhang J, Su A, Zhao L, Chen H, Hu Q: Comparison of effects on colitis-associated tumorigenesis and gut microbiota in mice between Ophiocordyceps sinensis and Cordyceps militaris. Phytomedicine 2021, 90: 153653.
- Cui L, Chen L, Yang G, Li Y, Qiao Z, Liu Y, Meng Y, Zhou Y, Sun L: Structural characterization and immunomodulatory activity of a heterogalactan from Panax ginseng flowers. Food Res Int 2021, 140: 109859.
- Ye J, Li L, Hu Z: Exploring the Molecular Mechanism of Action of Yinchen Wuling Powder for the Treatment of Hyperlipidemia, Using Network Pharmacology, Molecular Docking, and Molecular Dynamics Simulation. Biomed Res Int 2021, 2021: 9965906.
- Ru J, Li P, Wang J, Zhou W, Li B, Huang C, Li P, Guo Z, Tao W, Yang Y: TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. Journal of cheminformatics 2014, 6(1): 13.
- Kim S, Chen J, Cheng T, Gindulyte A, He J, He S, Li Q, Shoemaker BA, Thiessen PA, Yu B: PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res 2021, 49(D1): D1388-D1395.
- Daina A, Michielin O, Zoete V: SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res 2019, 47(W1): W357-w364.
- Rebhan M, Chalifa-Caspi V, Prilusky J, Lancet D: GeneCards: integrating information about genes, proteins and diseases. Trends Genetics 1997, 13(4): 163-163.
- Amberger JS, Bocchini CA, Schiettecatte F, Scott AF, Hamosh A: OMIM. org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res 2015, 43(D1): D789-D798.
- Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, Doncheva NT, Legeay M, Fang T, Bork P: The STRING database in 2021: customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res2021, 49(D1): D605-D612.
- Huang DW, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC: DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res 2007, 35(suppl_2): W169-W175.
- Rose Y, Duarte JM, Lowe R, Segura J, Bi C, Bhikadiya C, Chen L, Rose AS, Bittrich S, Burley SK: RCSB Protein Data Bank: architectural advances towards integrated searching and efficient access to macromolecular structure data from the PDB archive. J Mol Biol 2021, 433(11): 166704.
- Soudy M, Anwar AM, Ahmed EA, Osama A, Ezzeldin S, Mahgoub S, Magdeldin S: UniprotR: Retrieving and visualizing protein sequence and functional information from Universal Protein Resource (UniProt knowledgebase). J Proteomics 2020, 213: 103613.
- Tian C, Kasavajhala K, Belfon KA, Raguette L, Huang H, Migues AN, Bickel J, Wang Y, Pincay J, Wu Q: ff19SB: amino-acid-specific protein backbone parameters trained against quantum mechanics energy surfaces in solution. J Chem Theory Comput 2019, 16(1): 528-552.
- Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA: Development and testing of a general amber force field. J Comput Chem 2004, 25(9): 1157-1174.
- Roe DR, Cheatham III TE: PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J Chem Theory Comput 2013, 9(7): 3084-3095.
- Miller III BR, McGee Jr TD, Swails JM, Homeyer N, Gohlke H, Roitberg AE: MMPBSA. py: an efficient program for end-state free energy calculations. J Chem Theory Comput 2012, 8(9): 3314-3321.
- Malik S, Fatima B, Hussain D, Imran M, Chohan TA, Khan MS, Majeed S, Najam-ul-Haq M: Synthesis of novel nonsteroidal anti-inflammatory galloyl β-sitosterol-loaded lignin-capped Ag-based drug. Inflammopharmacology 2024, 32(2): 1333-1351.
- Zhang P, Liu N, Xue M, Zhang M, Liu W, Xu C, Fan Y, Meng Y, Zhang Q, Zhou Y: Anti-inflammatory and antioxidant properties of β-sitosterol in copper sulfate-induced inflammation in zebrafish (Danio rerio). Antioxidants 2023, 12(2): 391.
- Jiang Y, Liu Z, Hu J, Sun S, Xie X, Kong X, Tang Y: Pharmacodynamic evaluation of Cordyceps sinensis (Berk.) Sacc. for ischemic stroke in rats and potential mechanism through network pharmacology and molecular docking. J Trad Chin Med Sci 2023, 10(2): 196-207.
- Huang J, Lin Z, Wang Y, Ding X, Zhang B: Wuling san based on network pharmacology and in vivo evidence against hyperuricemia via improving oxidative stress and inhibiting inflammation. Drug Des Devel Thery 2023, Epub ahead of print.: 675-690.
- Yang S, Zhang Y, Zheng C: β-sitosterol mitigates apoptosis, oxidative stress and inflammatory response by inactivating TLR4/NF-кB pathway in cell models of diabetic nephropathy. Cell Biochem and Biophys 2025, 83(1): 1249-1262.
- Zhang F, Liu Z, He X, Li Z, Shi B, Cai F: β-Sitosterol-loaded solid lipid nanoparticles ameliorate complete Freund’s adjuvant-induced arthritis in rats: involvement of NF-кB and HO-1/Nrf-2 pathway. Drug deliv 2020, 27(1): 1329-1341.
- Jayaraman S, Devarajan N, Rajagopal P, Babu S, Ganesan SK, Veeraraghavan VP, Palanisamy CP, Cui B, Periyasamy V, Chandrasekar K: β-Sitosterol circumvents obesity induced inflammation and insulin resistance by down-regulating IKKβ/NF-κB and JNK signaling pathway in adipocytes of type 2 diabetic rats. Molecules 2021, 26(7): 2101.
- Jayaraman S, Prasad M, Natarajan SR, Krishnamoorthy R, Alshuniaber MA, Gatasheh MK, Veeraraghavan VP, Rajagopal P, Palanisamy CP: Molecular mechanisms underlying the effects of beta-sitosterol on TGF-β1/Nrf2/SIRT1/p53-mediated signaling in the kidney of a high-fat diet and sucrose-induced type-2 diabetic rat. Chem Biol Interact 2025, 411: 111443.
- Pitchot W, Hansenne M, Pinto E, Reggers J, Fuchs S, Ansseau M: 5-Hydroxytryptamine 1A receptors, major depression, and suicidal behavior. Biol psychiatry 2005, 58(11): 854-858.
- Wei S-Z, Yao X-Y, Wang C-T, Dong A-Q, Li D, Zhang Y-T, Ren C, Zhang J-B, Mao C-J, Wang F: Pramipexole regulates depression-like behavior via dopamine D3 receptor in a mouse model of Parkinson’s disease. Brain Res Bull 2021, 177: 363-372.
- Poyourow CN, Leonberg K, Ghajar M, Chung M, Byham-Gray L: The role of dietary acid load on progression of estimated glomerular filtration rate among individuals diagnosed with chronic kidney disease. J Ren Nutr 2024, 34(4): 273-282.
- van Baar MJ, van Bommel EJ, Smits MM, Touw DJ, Nieuwdorp M, Ten Kate RW, Joles JA, van Raalte DH: Whole-body insulin clearance in people with type 2 diabetes and normal kidney function: Relationship with glomerular filtration rate, renal plasma flow, and insulin sensitivity. J Diabetes Complications 2022, 36(4): 108166.
- Ruhé HG, Dekker JJ, Peen J, Holman R, De Jonghe F: Clinical use of the Hamilton depression rating scale: is increased efficiency possible? A post hoc comparison of Hamilton depression rating scale, Maier and Bech subscales, clinical global impression, and symptom Checklist-90 scores. Compr psychiatry 2005, 46(6): 417-427.
- Wang X, Bakar MHA, Kassim MA, Shariff KA, Rosdi MNM: Renoprotective mechanisms of Celastrol in high glucose-mediated HK-2 cell injury through Inhibition of the PI3K/Akt/NF-κB signalling pathway. Biochem Biophys Rep 2025, 41: 101928.
- Gong P, Long H, Yang Q, Zhou R, Yang W, Chen F, Xie J, Zhao Y, Xu H: Astragaloside IV alleviates diabetic nephropathy by modulating the gut-kidney axis and AMPK/PI3K/AKT pathway. Food Biosci 2024, 62: 105448.
- Zhou W-J, Liang W, Hu M-X, Ma Y-K, Yu S, Jin C, Li J-Q, Wang C, Wang C-Z, Gong P: Qingshen granules inhibits dendritic cell glycolipid metabolism to alleviate renal fibrosis via PI3K-AKT-mTOR pathway. Phytomedicine 2024, 135: 156148.
- Chen M, Chen Y, Zhu W, Yan X, Xiao J, Zhang P, Liu P, Li P: Advances in the pharmacological study of Chinese herbal medicine to alleviate diabetic nephropathy by improving mitochondrial oxidative stress. Biomed Pharmacother 2023, 165: 115088.
- Ye Y, Li M, Chen W, Wang H, He X, Liu N, Guo Z, Zheng C: Natural polysaccharides as promising reno-protective agents for the treatment of various kidney injury. Pharmacol Res 2024, 207: 107301.
- Zeng J, Xie Z, Chen L, Peng X, Luan F, Hu J, Xie H, Liu R, Zeng N: Rosmarinic acid alleviate CORT-induced depressive-like behavior by promoting neurogenesis and regulating BDNF/TrkB/PI3K signaling axis. Biomed Pharmacother 2024, 170: 115994.
- Batt A, Singh K, Gupta JK, Chanchal DK, Kumar K, Dubey A, Kumar S, Jain D: A comprehensive review of cellular stress response pathway system of Rhizoma coptidis. Pharmacol Res-Modern Chin Med 2024, 12: 100491.
- He W-W, Zeng X-X, Qi X-L, Gui C-Z, Liao W, Tu X, Deng J, Dong Y-T, Hong W, He Y: Regulating effect of miR-132–3p on the changes of MAPK pathway in rat brains and SH-SY5Y cells exposed to excessive fluoride by targeting expression of MAPK1. Ecotoxicol Environ Saf 2024, 279: 116467.
- Gong X-x, Cao L-h, Ni H-x, Zang Z-y, Chang H: Chinese herbal medicine for the treatment of diabetic nephropathy: from clinical evidence to potential mechanisms. J Ethnopharmacol 2024, 330: 118179.
- Utpal BK, Sutradhar B, Zehravi M, Sweilam SH, Durgawale TP, Arjun UVNV, Shanmugarajan TS, Kannan SP, Prasad PD, Usman MRM: Cellular stress response and neuroprotection of flavonoids in neurodegenerative diseases: Clinical insights into targeted therapy and molecular signaling pathways. Brain Res 2025, 1847: 149310.
- Hauger RL, Saelzler UG, Pagadala MS, Panizzon MS: The role of testosterone, the androgen receptor, and hypothalamic-pituitary–gonadal axis in depression in ageing Men. Rev Endocr Metab Disord 2022, 23(6): 1259-1273.
- Palimeri S, Palioura E, Piperi C, Kandaraki E, Sergentanis T, Levidou G, Papalois A, Korkolopoulou P, Papavassiliou AG, Diamanti-Kandarakis E: Additive effects of dietary glycotoxins and androgen excess on the kidney of a female rat model. Alexand J Med 2016, 52(2): 159-168.
- Yao W, Tao R, Xu Y, Chen Z-S, Ding X, Wan L: AR/RKIP pathway mediates the inhibitory effects of icariin on renal fibrosis and endothelial-to-mesenchymal transition in type 2 diabetic nephropathy. J Ethnopharmacol 2024, 320: 117414.
- Peltier MR, Verplaetse TL, Mineur YS, Gueorguieva R, Petrakis I, Cosgrove KP, Picciotto MR, McKee SA: Sex differences in progestogen-and androgen-derived neurosteroids in vulnerability to alcohol and stress-related disorders. Neuropharmacology 2021, 187: 108499.
- Yin Y, Ju T, Zeng D, Duan F, Zhu Y, Liu J, Li Y, Lu W: “Inflamed” depression: A review of the interactions between depression and inflammation and current anti-inflammatory strategies for depression. Pharmacol Res 2024, 207: 107322.
- Wojnicz A, Ortiz JA, Casas AI, Freitas AE, Lopez MG, Ruiz-Nuno A: Simultaneous determination of 8 neurotransmitters and their metabolite levels in rat brain using liquid chromatography in tandem with mass spectrometry: Application to the murine Nrf2 model of depression. Clin Chim Acta 2016, 453: 174-181.
Asia-Pacific Journal of Pharmacotherapy & Toxicology
p-ISSN: 2788-6840
e-ISSN: 2788-6859
 Copyright © Asia Pac J Pharmacother Toxicol. This work is licensed under a Creative Commons Attribution-NonCommercial-No Derivatives 4.0 International (CC BY-NC-ND 4.0) License.
Copyright © Asia Pac J Pharmacother Toxicol. This work is licensed under a Creative Commons Attribution-NonCommercial-No Derivatives 4.0 International (CC BY-NC-ND 4.0) License.

 Submit Manuscript
Submit Manuscript