Review Article | Open Access
Prospects of universal influenza virus vaccine and the current challenges of new antiviral drugs
Samer K Almadar1, Nasser M Alqahtani1
1Department of Medical Laboratory, College of Applied Medical Sciences, Prince Sattam Bin Abdulaziz University, Al-Kharj 11942, Saudi Arabia.
Correspondence: Nasser M Alqahtani (Department of Medical Laboratory, College of Applied Medical Sciences, Prince Sattam Bin Abdulaziz University, Al-Kharj 11942, Saudi Arabia; E-mail: qn832062@gmail.com).
Asia-Pacific Journal of Pharmacotherapy & Toxicology 2024, 4: 55-64. https://doi.org/10.32948/ajpt.2024.09.25
Received: 20 Sep 2024 | Accepted: 28 Sep 2024 | Published online: 09 Oct 2024
Key words innate immunity, influenza virus, universal vaccine, neutralizing antibodies
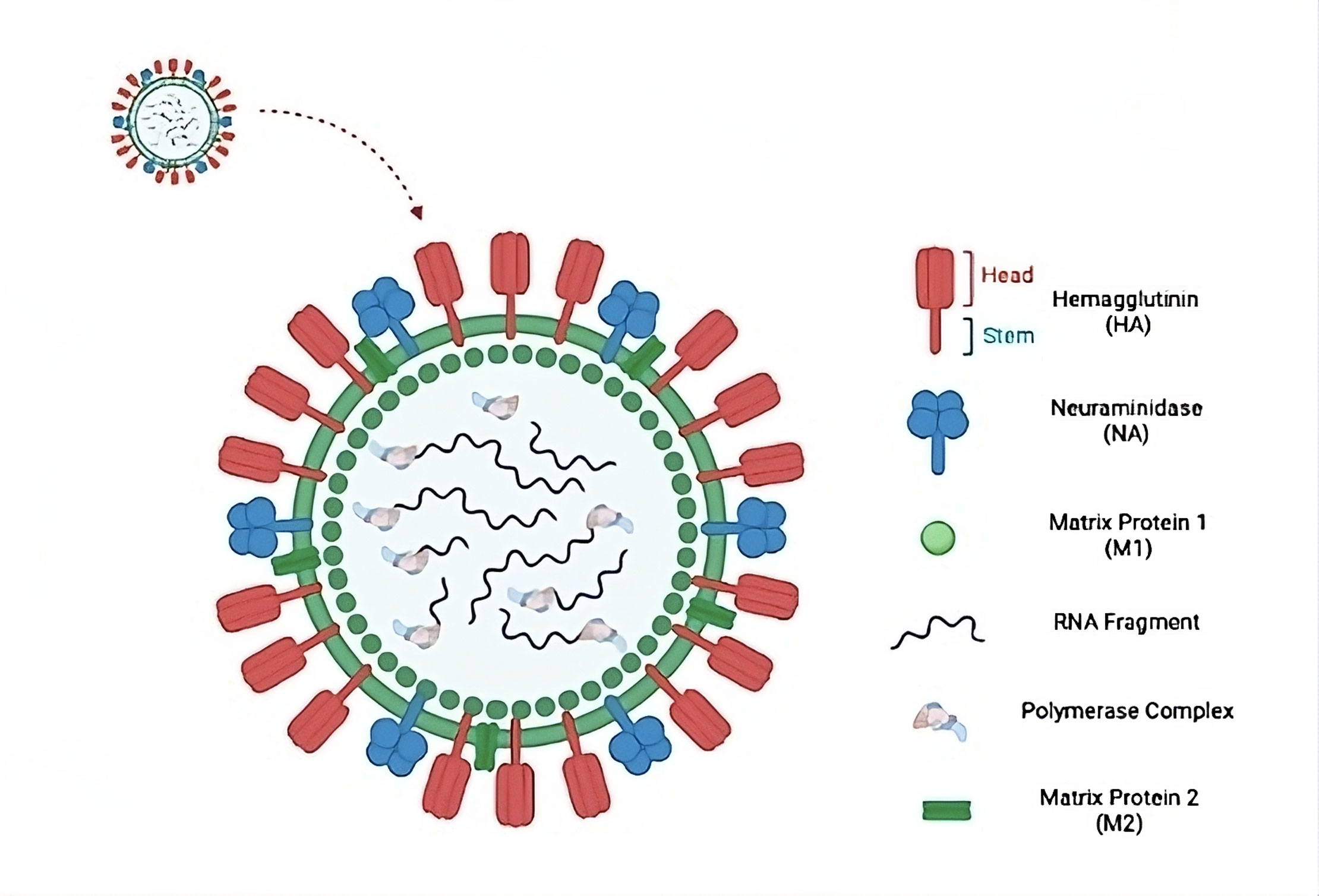 Figure 1. Influenza A virus and its components structural overview. (HA): a glycoprotein on the surface that helps the virus attach to host cells and make entrance easier. (NA): a surface enzyme that facilitates the release of new virions by cleaving the sialic acid present on the host cell. (M1): a structural protein that maintains the structural integrity and aids in viral assembly on the inner surface of the viral membrane. (RNA fragment): viral variety and genetic reassortment are made possible by the segmented RNA genome that codes for viral proteins. (Polymerase complex): a multi-subunit complex that is in charge of the viral RNA genome's transcription and replication. (M2): an ion channel that permits the release of viral RNA by enabling viral uncoating within host cells.
Figure 1. Influenza A virus and its components structural overview. (HA): a glycoprotein on the surface that helps the virus attach to host cells and make entrance easier. (NA): a surface enzyme that facilitates the release of new virions by cleaving the sialic acid present on the host cell. (M1): a structural protein that maintains the structural integrity and aids in viral assembly on the inner surface of the viral membrane. (RNA fragment): viral variety and genetic reassortment are made possible by the segmented RNA genome that codes for viral proteins. (Polymerase complex): a multi-subunit complex that is in charge of the viral RNA genome's transcription and replication. (M2): an ion channel that permits the release of viral RNA by enabling viral uncoating within host cells.
Selecting a vaccine strain that closely matches the circulating influenza strain is a complex process and often presents challenges, which may result in less-than-optimal protection. These challenges arise from the ongoing antigenic changes that occur in the HA molecule of the influenza virus, which can cause the virus to evade recognition by the immune response. Additionally, the emergence of novel antigenic variants cannot always be accurately predicted, further complicating the selection of an appropriate vaccine strain. According to the Centers for Disease Control (CDC), it is estimated that annually, the prevention of influenza B virus infections could potentially lead to the avoidance of 40,000 to 275,000 illnesses [7]. Hence, there is a dire need for alternative strategies to effectively control emerging influenza viruses.
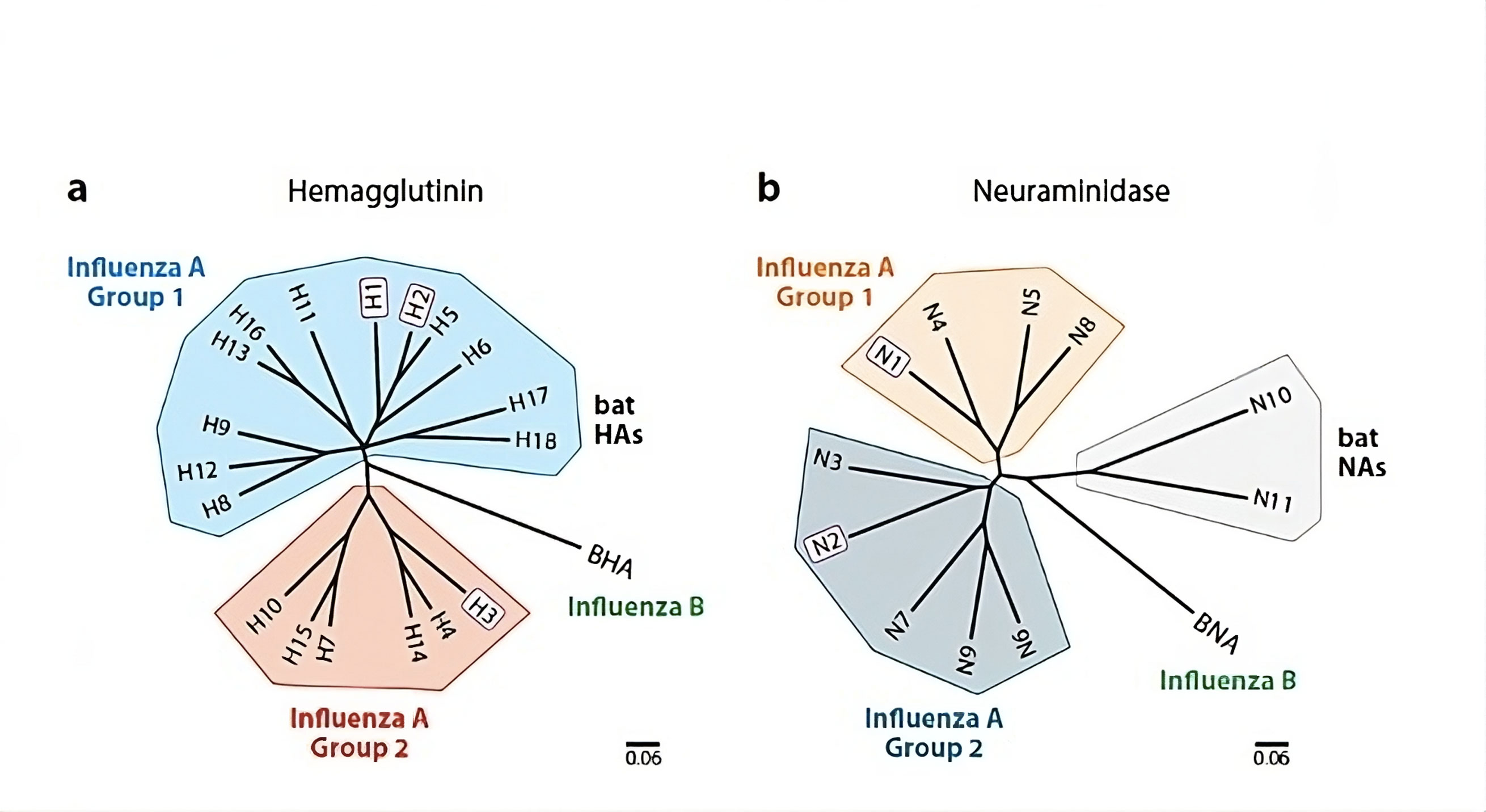 Figure 2. Phylogenetic trees illustrating the relationships of (a) hemagglutinin (HA) and (b) neuraminidase (NA) reveal the classification of these major surface glycoproteins of influenza viruses into distinct groups and subtypes. Notably, only those viruses featuring H1, H2, or H3 hemagglutinins and N1 or N2 neuraminidases—such as H1N1, H2N2, or H3N2 (highlighted in purple)—have circulated globally within the human population over the past century. The scale bars on the trees represent a 6% variation at the amino acid level, providing insight into the genetic diversity among these influenza virus strains.
Figure 2. Phylogenetic trees illustrating the relationships of (a) hemagglutinin (HA) and (b) neuraminidase (NA) reveal the classification of these major surface glycoproteins of influenza viruses into distinct groups and subtypes. Notably, only those viruses featuring H1, H2, or H3 hemagglutinins and N1 or N2 neuraminidases—such as H1N1, H2N2, or H3N2 (highlighted in purple)—have circulated globally within the human population over the past century. The scale bars on the trees represent a 6% variation at the amino acid level, providing insight into the genetic diversity among these influenza virus strains.
Structural analyses revealed that broadly neutralizing antibodies against influenza viruses, such as CR6261, F10, F16, CR8020, CR8033, and CR8071, target distinct sites on the stem region of the HA molecule. Some antibodies like CR8033 and CR8071 are capable of neutralizing influenza B viruses from both Yamagata and Victoria lineages by recognizing two distinct conserved epitopes on the HA of influenza B viruses. These antibodies inhibit virus progeny release by interfering with viral replication and preventing the release of new virus particles, thereby providing a potential mechanism for their antiviral activity [13]. Broadly neutralizing antibodies against influenza viruses are being developed as monoclonal antibody therapies in different countries [14]. Identifying vulnerabilities in the defense mechanisms of the influenza virus can also pave the way for the development of potential drug candidates, such as proteins and small molecules that mimic the interactions of antibodies, thereby competing for receptor binding [15]. Moreover, the subsequent objective is to develop a universal influenza vaccine that can trigger the production of antibodies targeting the conserved epitopes of the HA protein.
Universal vaccines can be designed by altering vaccine candidates to eliminate strain-specific, immunodominant epitopes located on the head region of the HA protein. Instead, these vaccines can incorporate cross-reactive, non-immunodominant epitopes found in the stem region. A key challenge in this strategy is improving the immune system's ability to access these conserved HA determinants. Although several headless HA immunogens have been developed by removing a significant portion of the HA1 region, the reasons for their failure to elicit a broadly cross-reactive neutralizing antibody feedback in animal models remain unclear [16-18].
For the past 60 years, the development of influenza vaccines has primarily focused on the highly variable HA-head region. However, further research and a deeper insight of the molecular mechanisms underlying neutralization of influenza viruses by both neutralizing and non-neutralizing antibodies are needed to generate an immune response. Recent data have also reported cases of vaccine-associated enhanced respiratory disease in pigs following vaccination with whole inactivated H1N2 (human-like) virus, and subsequent challenge with a different strain of H1N1pdm09 virus [19]. The authors of the study revealed that vaccination with whole inactivated H1N2 virus induced antibodies that cross-reacted with the H1N1pdm09 virus. As a result, this led to emphasized fusion of the H1N1pdm09 virus with target cells, ultimately resulting in increased disease severity. In addition to the mechanism mentioned above, there are two other potential ways in which antibodies may enhance disease: (1) non-neutralizing antibodies that specifically target the HA stem region may bind to the HA protein and direct the virus to cells that express Fc receptors. (2) Antibodies that target HA on the surface of influenza virus-infected cells may bind and trigger antibody-mediated complement activation, leading to inflammation and cell lysis [20]. Consequently, further research is needed to better understand the rational model of safe and effective universal influenza vaccines. This includes investigating ways that can provide durable protection against diverse influenza virus strains.
NAIs are a class of antiviral drugs that specifically target the enzymatically active site of the neuraminidase (NA) protein in influenza A and B viruses. By binding to this site, NAIs interfere with the protein's function, particularly its role in cleaving terminal sialic acid residues from the host cell surface, which is essential for the release of newly produced virions [21]. Consequently, NAIs prevent the release of budding viral particles from infected host cells, limiting the virus's spread to uninfected cells. Unlike antibiotics, which can effectively eliminate or significantly reduce bacterial presence, NAIs provide temporary relief by stopping the infection of new host cells with influenza viruses. For this reason, administering NAIs within 48 hours of symptom onset is critical for effective influenza treatment, which can be challenging in many regions [22]. However, some evidence indicates that NAIs may still offer benefits if given later in the disease, particularly in underdeveloped areas or for hospitalized patients. Administering NAIs early in the course of the infection can help modulate disease progression, allowing the immune system to clear the virus more rapidly. Despite this, the limited availability of only one class of antiviral drugs and the short therapeutic window for NAIs pose considerable challenges for healthcare providers.
Neuraminidase inhibitors
Of the four existing neuraminidase inhibitors, two have been widely used globally since the 1999-2000 flu season: oral oseltamivir (Tamiflu®, Roche) and inhaled zanamivir (Relenza®, GlaxoSmithKline). Additionally, intravenous peramivir (Rapiacta®, Peramiflu®, BioCryst Pharmaceuticals) has received approval for treating influenza in adults and children in Japan, South Korea, and China. Inhaled laninamivir octanoate (Inavir®, Daiichi Sankyo) is also approved in Japan for the treatment of influenza in both adults and children [23]. Peramivir has distinct structural features compared to oseltamivir and zanamivir, showing prolonged binding to the neuraminidase site for over 24 hours and slower dissociation. On the other hand, laninamivir octanoate has been proven to have clinical efficacy against seasonal influenza A and B viruses, as well as the H1N1pdm09 virus [24]. The active form of laninamivir persists at a high concentration in the lungs for at least 5 days [25]. As a result, a single dose of laninamivir octanoate can effectively treat influenza by providing a significant impact.
Resistance to neuraminidase inhibitors
The emergence of influenza viruses resistant to neuraminidase inhibitors poses a serious challenge to the effectiveness of antiviral treatments. Amino acid changes linked to NAI resistance can weaken the binding interaction between the neuraminidase enzyme and the inhibitor, allowing the virus to evade NAI treatment [23]. Molecular markers of resistance have been found at both the catalytic and framework residues of the neuraminidase glycoprotein, varying for oseltamivir, zanamivir, and peramivir. However, to date, no resistance-related substitutions have been reported for laninamivir. Oseltamivir, being a hydrophobic drug, requires rearrangements in the influenza NA protein to enable its binding. The E276 residue in NA must rotate and interact with R224 to create a receptive pocket for oseltamivir binding [26]. Any mutation that inhibits this rearrangement, such as preventing the rotation of E276 residue and the formation of the pocket, leading to a decrease in its efficacy. Amino acid substitutions associated with oseltamivir resistance in NA can vary depending on the NA types and subtypes of influenza A and B viruses. These amino acid changes can impair the binding of oseltamivir to the NA enzyme, leading to reduced efficacy of the drug against these resistant variants of influenza viruses [27].
NAI-resistant viruses can develop either due to drug-selection pressure or through natural processes without drug exposure. Before 2007, the rate of oseltamivir resistance in clinical trial samples was reported as 0.3% in adults and 4% in children [28, 29]. However, during the 2007-2009 influenza seasons, oseltamivir-resistant A/Brisbane/59/2007 (H1N1)-like viruses (subclade 2B) with the NA H274Y resistance marker became widespread globally. Despite this, epidemiological data did not link the emergence of resistance directly to oseltamivir usage. This suggests that the H274Y mutation in the NA protein arose naturally, leading to viruses with enhanced transmissibility and fitness compared to their drug-sensitive counterparts [30-32].
Different changes in the NA protein can mitigate the negative effects of the H274Y mutation, such as the D344N, Q222R, and V234M substitutions, which help restore NA affinity and surface accumulation [33-35]. Such changes allow the H274Y mutation to persist without compromising viral function. Since the 2009 pandemic's first wave and the subsequent disappearance of the A/Brisbane/59/2007 (H1N1) strain, oseltamivir resistance in H1N1pdm09 variants has remained low, around 1% [36, 37]. Nonetheless, Vietnam, Australia, and the UK have reported clusters of oseltamivir-resistant infections that are not associated with therapy and most likely involve the spread of H1N1pdm09 mutant strains [38-40]. The intricacy of NAI-resistance patterns draws attention to the difficulties in conducting antiviral monitoring, which is essential for determining the hazards associated with newly emerging viruses, particularly those originating from avian or human origins.
Development of novel antiviral drugs with multiple pathway targeting
The heavy reliance on NAIs carries the same risk as any monotherapy: the potential for drug resistance. The rise and global spread of naturally occurring NAI-resistant H1N1 influenza viruses during the 2007-2009 seasons underscore the pressing need for new antiviral options and combination therapies. Future treatments could focus on directly inhibiting viral proteins or essential cellular components for viral replication, or by modulating the host's immune response. Several drugs in development are aimed at targets like the viral polymerase (e.g., Favipiravir), hemagglutinin (e.g., nitazoxanide), or sialic acid receptors (e.g., Fludase). Additionally, anti-inflammatory compounds that do not promote antiviral resistance could play a crucial role in improving outcomes in severe influenza cases, either as standalone treatments or part of a combination strategy. Other promising approaches include enhancing the host’s innate immune response or employing immunotherapy using antibodies targeting conserved regions on the hemagglutinin protein. Diversifying treatment options and exploring combination therapies can be essential in mitigating the risk of drug resistance and improving outcomes in the management of influenza infections.
CD8+ CTLs are capable of recognizing a wide array of influenza virus antigens, including conserved internal viral proteins presented by HLA class molecules [45, 46]. This ability allows the cellular immune system to mount cross-reactive responses against various influenza strains due to its recognition of these conserved proteins [47-49]. Conversely, though the humoral immune feedback primarily functions by producing antibodies that target viral surface proteins like HA, NA, and to a lesser extent, matrix protein 2 (M2), which is less prevalent on the virus surface. HA, the main antigenic protein, plays a key role in viral attachment and membrane fusion. Therefore, antibodies directed against HA can neutralize the virus and prevent infection.
HA is a homotrimer; each monomer is made up of the two domains, HA1 and HA2, which are joined by a disulfide link. Whereas HA1 creates the distal globular head, which houses the majority of the antigenic areas and receptor-binding sites, HA2 forms the membrane anchor and the "stem" region. As mentioned earlier, antigenic drift in circulating strains primarily results from mutations in the antigenic sites of the HA1 globular head. Between 1999 and 2010, it was estimated that human seasonal H3 and H1 influenza viruses experienced amino acid changes in the range of 2.1% to 3% per drift variant [50]. In contrast, the HA2 region remained highly conserved, with only three amino acid changes observed in both H1 and H3 strains during the same period. Despite its conservation, HA2 is immunogenic, and antibodies targeting this region have been detected in humans following infection and vaccination, though they are produced at much lower levels than those targeting the globular head [51, 52].
The second primary surface glycoprotein of the influenza virus, neuraminidase (NA), is an enzyme that helps release freshly produced viruses by cleaving sialic acid residues from the surface of the host cell. While antibodies targeting NA do not prevent initial infection, they can help limit the spread of the virus. Similarly, the viral proteins M2 and nucleoprotein (NP) are critical to the virus replication process. Although antibodies against M2 and NP do not neutralize the virus directly, studies in mice have shown that they can reduce viral replication and contribute to controlling the infection [53].
Hemagglutination inhibition or neutralization assays are commonly used to detect the induction of HA-specific antibodies in the blood, which is a crucial indicator of recent influenza exposure or vaccination. For inactivated influenza vaccines in adults, a titer of ≥40 is often considered an immune correlate of protection based on findings from vaccine trials and clinical studies. However, for other vaccine platforms, established immune correlates of protection are lacking, which has been recognized as a significant barrier to advancing new influenza vaccine strategies [54]. Assessing the direct protective efficacy of a vaccine its ability to prevent infection in real-world settings presents additional challenges. These include the need for large study populations and the capacity to track participants across at least one flu season, making empirical evaluation difficult [55]. As a result, the process of developing a clinically validated influenza vaccine is both time-consuming and expensive.
No applicable.
Ethics approval
No applicable.
Data availability
The data will be available upon request.
Funding
None.
Authors’ contribution
Samer K Almadar and Nasser M Alqahtani contributed to draft, critical revision of the article. Nasser M Alqahtani approved the final version to be submited.
Competing interests
None.
- Uhart M, Bricout H, Clay E, Largeron N: Public health and economic impact of seasonal influenza vaccination with quadrivalent influenza vaccines compared to trivalent influenza vaccines in Europe. Hum Vaccines Immunother 2016, 12(9): 2259-2268.
- Molinari NAM, Ortega-Sanchez IR: Messonnier ML, Thompson WW, Wortley PM, Weintraub E, Bridges CB: The annual impact of seasonal influenza in the US: measuring disease burden and costs. Vaccine 2007, 25(27): 5086-5096.
- Preaud E, Durand L, Macabeo B, Farkas N, Sloesen B, Palache A, Shupo F, Samson S I: Annual public health and economic benefits of seasonal influenza vaccination: a European estimate. BMC Public Health 2014, 14(1): 1-12.
- Petrie JG, Ohmit SE, Johnson E, Truscon R, Monto AS: Persistence of antibodies to influenza hemagglutinin and neuraminidase following one or two years of influenza vaccination. J Infect Dis 2015, 212(12): 1914-1922.
- Chiatti C, Barbadoro P, Lamura G, Di Stanislao F, Prospero E: Improving the delivery of flu vaccine for the older people in times of economic recession: what social epidemiology tells us, and what else we need to know. Human vaccines 2011, 7(9): 986-988.
- Fiore AE, Uyeki TM, Broder K, Finelli L, Euler GL, Singleton JA, Iskander JK, Wortley PM, Shay DK, Bresee JS: Prevention and control of influenza with vaccines: recommendations of the Advisory Committee on Immunization Practices (ACIP), 2010. MMWR Recomm Rep 2010, 59(RR-8): 1-62.
- Reed C, Meltzer MI, Finelli L, Fiore A: Public health impact of including two lineages of influenza B in a quadrivalent seasonal influenza vaccine. Vaccine 2012, 30(11): 1993-1998.
- Okuno Y, Isegawa Y, Sasao F, Ueda S: A common neutralizing epitope conserved between the hemagglutinins of influenza A virus H1 and H2 strains. J Virol 1993, 67(5): 2552-2558.
- Friesen RH, Lee PS, Stoop EJ, Hoffman RM, Ekiert DC, Bhabha G: A common solution to group 2 influenza virus neutralization. Proc Natl Acad Sci 2014, 111(1): 445-450.
- Yoshida R, Igarashi M, Ozaki H, Kishida N, Tomabechi D, Kida H, Ito K, Takada A: Cross-protective potential of a novel monoclonal antibody directed against antigenic site B of the hemagglutinin of influenza A viruses. PLoS Pathog 2009, 5(3): e1000350.
- Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D: A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 2011, 333(6044): 850-856.
- Dreyfus C, Laursen NS, Kwaks T, Zuijdgeest D, Khayat R: Highly conserved protective epitopes on influenza B viruses. Science 2012, 337(6100): 1343-1348.
- Sun X, Ling Z, Yang Z, Sun B: Broad neutralizing antibody-based strategies to tackle influenza. Curr Opin Virol 2022, 53: 101207.
- Trombetta CM, Kistner O, Montomoli E, Viviani S, Marchi S: Influenza Viruses and Vaccines: The Role of Vaccine Effectiveness Studies for Evaluation of the Benefits of Influenza Vaccines. Vaccines 2022, 10(5): 714.
- Xu R, Krause JC, McBride R, Paulson JC, Crowe JE, Wilson IA: A recurring motif for antibody recognition of the receptor-binding site of influenza hemagglutinin. Nat Struc Mol Biol 2013, 20(3): 363-370.
- Mintaev R,Glazkova D, Bogoslovskaya E, Shipulin G: Immunogenic epitope prediction to create a universal influenza vaccine. Heliyon 2022, 8(5): e09364.
- Allen JD, Ross TM: Next generation methodology for updating HA vaccines against emerging human seasonal influenza A (H3N2) viruses. Sci Rep 2021, 11(1): 1-14.
- Wang TT, Tan GS, Hai R, Pica N, Ngai L, Ekiert DC, Wilson IA, García-Sastre A, Moran TM, Palese P: Vaccination with a synthetic peptide from the influenza virus hemagglutinin provides protection against distinct viral subtypes. Proc Natl Acad Sci 2010, 107(44): 18979-18984.
- Khurana S, Loving CL, Manischewitz J, King LR, Gauger PC, Henningson J, Vincent, AL, Golding H: Vaccine-induced anti-HA2 antibodies promote virus fusion and enhance influenza virus respiratory disease. Sci Transl Med 2013, 5(200): 200ra114.
- Krammer F: The quest for a universal flu vaccine: headless HA 2.0. Cell host microbe 2015, 18(4): 395-397.
- Air GM: Influenza neuraminidase. Influenza Other Respir. Viruses 2012, 6(4): 245-256.
- Aoki FY, Macleod MD, Paggiaro P, Carewicz O, El Sawy A, Wat C, Griffiths M, Waalberg E, Ward P: Early administration of oral oseltamivir increases the benefits of influenza treatment. J Antimicrob Chemother 2003, 51(1): 123-129.
- Wathen MW, Barro M, Bright RA: Antivirals in seasonal and pandemic influenza-future perspectives. Influenza Other Respir. Viruses 2013, 7 Suppl 1(Suppl 1): 76-80.
- Bantia S, Arnold CS, Parker CD, Upshaw R, Chand P: Anti-influenza virus activity of peramivir in mice with single intramuscular injection. Antivir Res 2006, 69(1): 39-45.
- Ikematsu H, Kawai N: Laninamivir octanoate: a new long-acting neuraminidase inhibitor for the treatment of influenza. Expert Rev. Anti Infect Ther 2011, 9(10): 851-857.
- Collins PJ, Haire LF, Lin YP, Liu J, Russell RJ, Walker PA Skehel JJ, Martin SR, Hay AJ, Gamblin SJ et al: Crystal structures of oseltamivir-resistant influenza virus neuraminidase mutants. Nature 2008, 453(7199): 1258-1261.
- Samson M, Pizzorno A, Abed Y, Boivin G: Influenza virus resistance to neuraminidase inhibitors. Antivir Res 2013, 98(2): 174-185.
- Monto AS, McKimm-Breschkin JL, Macken C, Hampson AW, Hay A, Klimov A, Tashiro M, Webster RG, Aymard M, Hayden FG et al: Detection of influenza viruses resistant to neuraminidase inhibitors in global surveillance during the first 3 years of their use. Antimicrob Agents Chemother 2006, 50(7): 2395-2402.
- Ward P, Small I, Smith J, Suter P, Dutkowski R: Oseltamivir (Tamiflu®) and its potential for use in the event of an influenza pandemic. J Antimicrob Chemother 2005, 55 Suppl 1: i5-i21.
- Dharan NJ, Gubareva LV, Meyer JJ, Okomo-Adhiambo M, McClinton RC, Marshall SA, George KS, Epperson S, Brammer L, Klimov AI et al: Infections with oseltamivir-resistant influenza A (H1N1) virus in the United States. Jama 2009, 301(10): 1034-1041.
- Hauge SH, Dudman S, Borgen K, Lackenby A, Hungnes O: Oseltamivir-resistant influenza viruses A (H1N1), Norway, 2007-08. Emerg Infect Dis 2009, 15(2): 155-162.
- Meijer A, Lackenby A, Hungnes O, Lina B, Van Der Werf S, Schweiger B, Opp M, Paget J, Van De Kassteele J, Hay A et al: Oseltamivir-resistant influenza virus A (H1N1), Europe, 2007-08 season. Emerg. Infect Dis 2009, 15(4): 552-560.
- Collins P, Haire L, Lin Y, Liu J, Russell R, Walker P, Martin S, Daniels R, Gregory V, Skehel J et al: Structural basis for oseltamivir resistance of influenza viruses. Vaccine 2009, 27(45): 6317-6323.
- Rameix-Welti MA, Munier S, Le Gal S, Cuvelier F, Agou F, Enouf V, Naffakh N, Van Der Werf S: Short communication Neuraminidase of 2007–2008 influenza A (H1N1) viruses shows increased affinity for sialic acids due to the D344N substitution. Antivir Ther 2011, 16(4), 597-603.
- Bloom JD, Gong LI, Baltimore D: Permissive secondary mutations enable the evolution of influenza oseltamivir resistance. Science 2010, 328(5983): 1272-1275.
- Renaud C, Kuypers J, Englund JA: Emerging oseltamivir resistance in seasonal and pandemic influenza A/H1N1. J Clin Virol 2011, 52(2): 70-78.
- Storms AD, Gubareva LV, Su S, Wheeling JT, Okomo-Adhiambo M, Pan CY, Reisdorf E, George KS, Myers R, Wotton JT et al: Oseltamivir-resistant pandemic (H1N1) 2009 virus infections, United States, 2010–11. Emerg Infect Dis 2012, 18(2): 308-311.
- Lackenby A, Gilad JM, Pebody R, Miah S, Calatayud L, Bolotin S, Vipond I, Muir P, Guiver M, McMenamin J et al: Continued emergence and changing epidemiology of oseltamivir-resistant influenza A (H1N1) 2009 virus, United Kingdom, winter 2010/11. Eurosurveillance 2011, 16(5): 19784.
- Hurt AC, Hardie K, Wilson NJ, Deng YM, Studies MO, BN Grad Dip Legal, Gehrig N, Kelso A: Community transmission of oseltamivir-resistant A (H1N1) pdm09 influenza. N Engl J Med 2011, 365(26): 2541-2542.
- Mai LQ, Wertheim HF, Duong TN, van Doorn HR, Hien NT, Horby PA: A community cluster of oseltamivir-resistant cases of 2009 H1N1 influenza. N Engl J Med 2010, 362(1): 86-87.
- Carrat F, Vergu E, Ferguson NM, Lemaitre M, Cauchemez S, Leach S, Valleron AJ: Time lines of infection and disease in human influenza: a review of volunteer challenge studies. Am J Epidemiol 2008, 167(7): 775-785.
- Pang IK, Iwasaki A: Inflammasomes as mediators of immunity against influenza virus. Trends Iimmunol 2011, 32(1): 34-41.
- Kreijtz J, Fouchier R, Rimmelzwaan, G: Immune responses to influenza virus infection. Virus Res 2011, 162(1-2): 19-30.
- Yap K, Ada G, McKenzie I: Transfer of specific cytotoxic T lymphocytes protects mice inoculated with influenza virus. Nature 1978, 273(5659): 238-239.
- Gianfrani C, Oseroff C, Sidney J, Chesnut RW, Sette A: Human memory CTL response specific for influenza A virus is broad and multispecific. Human Immunol 2000, 61(5): 438-452.
- Wahl A, Schafer F, Bardet W, Buchli R, Air GM, Hildebrand WH: HLA class I molecules consistently present internal influenza epitopes. Proc Natl Acad Sci 2009, 106(2): 540-545.
- Kees U, Krammer PH: Most influenza A virus-specific memory cytotoxic T lymphocytes react with antigenic epitopes associated with internal virus determinants. J Exp Med 1984, 159(2): 365-377.
- Ge X, Tan V, Bollyky PL, Standifer NE, James EA, Kwok WW: Assessment of seasonal influenza A virus-specific CD4 T-cell responses to 2009 pandemic H1N1 swine-origin influenza A virus. J Virol 2010, 84(7): 3312-3319.
- Babon J, AB, Cruz J, Ennis FA, Yin L, Terajima M: A human CD4+ T cell epitope in the influenza hemagglutinin is cross-reactive to influenza A virus subtypes and to influenza B virus. J Virol 2012, 86(17): 9233-9243.
- Qian M, Hu H, Zuo T, Wang G, Zhang L, Zhou P: Unraveling of a neutralization mechanism by two human antibodies against conserved epitopes in the globular head of H5 hemagglutinin. J Virol 2013, 87(6): 3571-3577.
- Varečková E, Kostolanský F, Mucha V: Monoclonal antibodies specific to the ha2 gp of influenza a virus haemagglutinin. Trends Monoclon Anti Res 2005, 123.
- Huang Y, França MS, Allen JD, Shi H, Ross TM: Next Generation of computationally optimized broadly reactive ha vaccines elicited cross-reactive immune responses and provided protection against H1N1 virus infection. Vaccines 2021, 9(7): 793.
- Mozdzanowska K, Maiese K, Furchner M, Gerhard W: Treatment of influenza virus-infected SCID mice with nonneutralizing antibodies specific for the transmembrane proteins matrix 2 and neuraminidase reduces the pulmonary virus titer but fails to clear the infection. Virology 1999, 254(1): 138-146.
- Hampson AW, Osterhaus AD, Pervikov Y, Kieny MP: Report of the second meeting on the development of influenza vaccines that induce broad-spectrum and long-lasting immune responses, World Health Organization, Geneva, Switzerland, 6-7 December 2005. Vaccine 2006, 24(23): 4897-4900.
- Zhang J, Huang L, Liao M, Qi W: Comparative efficacy of inactivated and live attenuated influenza vaccines. Lancet Microbe 2023, 4(2): e70-e71.
Asia-Pacific Journal of Pharmacotherapy & Toxicology
p-ISSN: 2788-6840
e-ISSN: 2788-6859
 Copyright © Asia Pac J Pharmacother Toxicol. This work is licensed under a Creative Commons Attribution-NonCommercial-No Derivatives 4.0 International (CC BY-NC-ND 4.0) License.
Copyright © Asia Pac J Pharmacother Toxicol. This work is licensed under a Creative Commons Attribution-NonCommercial-No Derivatives 4.0 International (CC BY-NC-ND 4.0) License.

 Submit Manuscript
Submit Manuscript